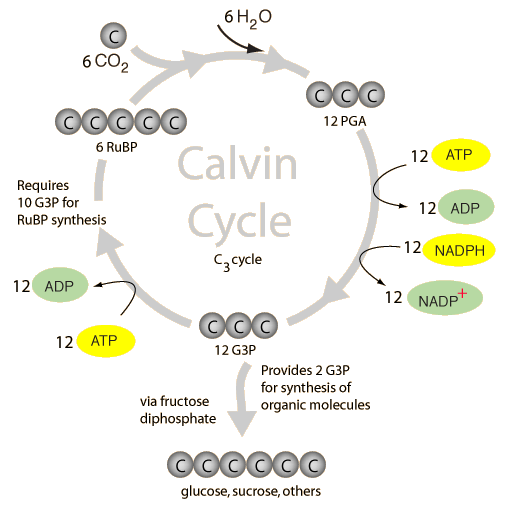
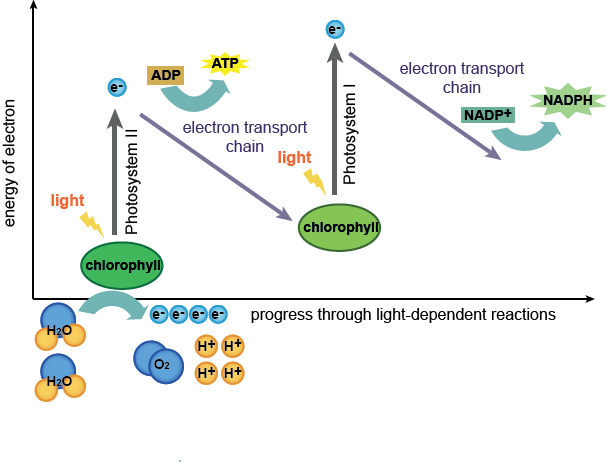
Image taken from: http://hyperphysics.phy-astr.gsu.edu/hbase/biology/imgbio/calvine.gif
- Happens in the stroma
- Does not require light
- ATP and NADPH from light dependent reaction is used to reduce carbon dioxide to triose phosphate and regenerate ribulose biphosphate (RuBP)
- Carbon dioxide reacts with RuBP to form an unstable 6-carbon intermediate which breaks down into two molecules of 3 phosphoglycerate (PGA) catalysed by ribulose biphosphate carboxylase/oxygenase (Rubisco)
- ATP and NADPH reduces PGA to form triose phosphate (G3P), forming ADP and NADP in the process
- Most of the triose phosphate is used to regenerate RuBP
- Some triose phosphate exit the cycle and is used to synthesise other products like starch and glucose
- ADP and NADP are channeled back into the light dependent reaction to produce more ATP and NADPH